The big takeaway:
While we know that gut microbes profoundly affect human health and well-being, there are still barriers to bringing microbiome diagnostics and therapeutics into clinical practice. In a new paper published in Gastroenterology, experts with the AGA Center for Gut Microbiome Research and Education outline the challenges in assessing the efficacy of microbiome-based therapeutics and recommend research that is needed to bring tests and therapies to patient care.
Here’s what experts recommend:
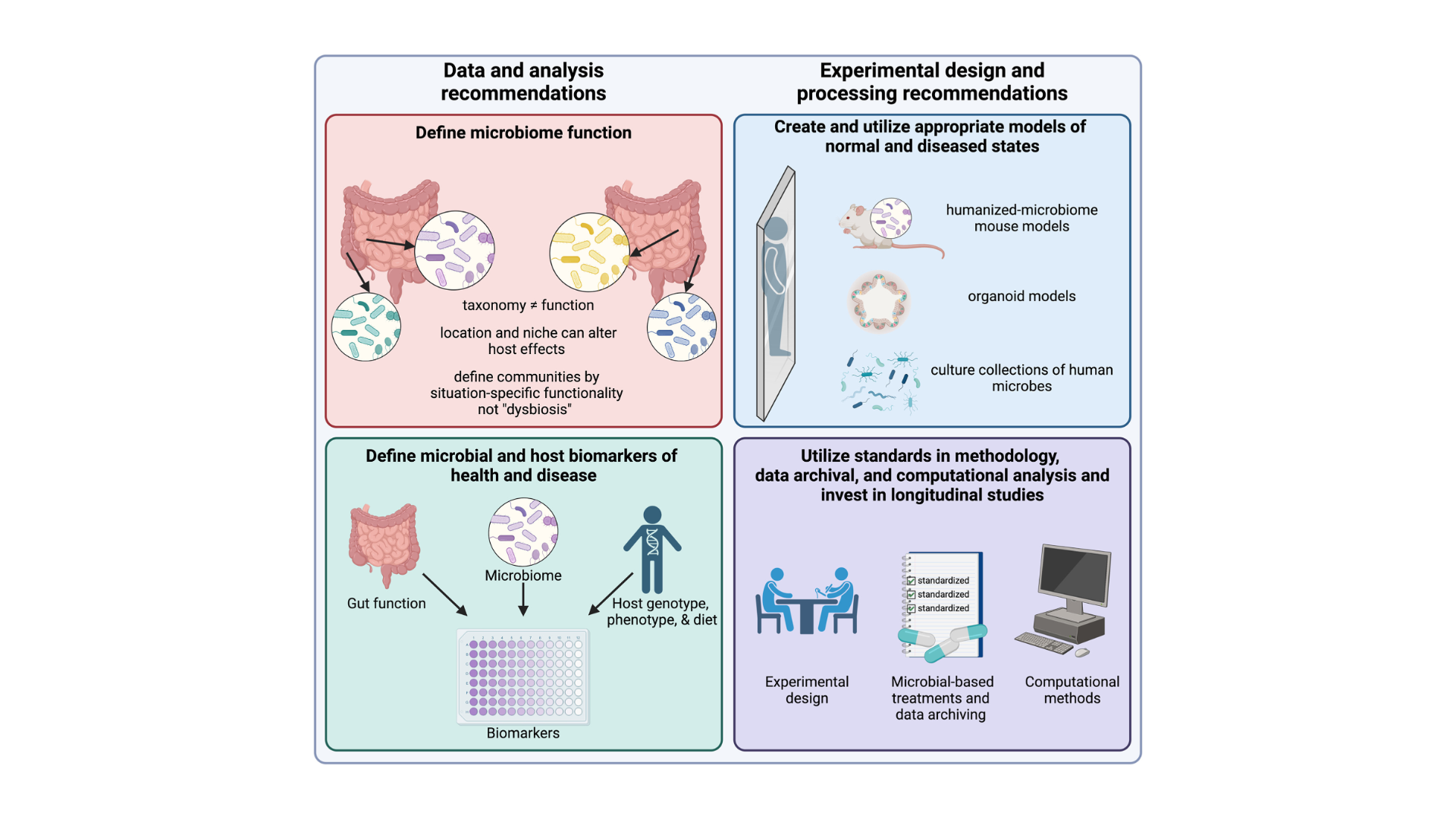
Easily defined biomarkers are needed to measure gut function and define intestinal dysfunction.
Fecal samples, a common proxy, fail to fully represent the distinct microbial communities within the entire gut. Better models are needed to address this to mimic human microbiome functionality and diseased states.
Advances in multi-omics and non-invasive sampling tools, like “smart pills,” could improve sampling accuracy.
Trials should pre-register protocols, use standardized methods, and deposit raw data in public repositories. The STORMS checklist is encouraged to ensure consistent study reporting and comparative analysis.
Researchers should prioritize understanding microbial functions and their mechanisms within the body over taxonomic classification.
Using established bioinformatics platforms for meta-analysis will help analyze microbiome data consistently.
It is critical to invest in studies with well-defined longitudinal cohorts and incorporate microbiome science into the collection of big data. Read the full paper in the November issue of Gastroenterology.